
Innovative Research
Providing state-of-the-art methodology for clinical, basic science and translational research empowers Duke Anesthesiology to explore revolutionary clinical inquiries by using innovative investigation methods.
Through significant research in neuroscience, molecular biology, molecular and human pharmacology endeavors, our team is making crucial advancements for patients worldwide.
Novel Adrb3 Antagonists for the Treatment of Chronic Pain
Andrea Nackley, PhD
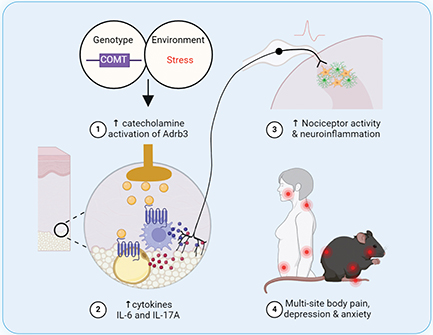
Chronic primary pain conditions (CPPCs), including fibromyalgia, headache, low back pain, and irritable bowel syndrome, affect one in every three Americans, predominantly females. CPPCs are characterized by pain lasting three months or longer in the absence of obvious injury or infection and they tend to co-occur. This co-occurring pain creates serious problems for patients, adding to the suffering and disability caused by a single pain condition. Health care providers who manage one type of chronic pain often are at a loss to manage pain occurring elsewhere in the body. All too often, patients are referred from one specialist to another at great expense and little relief. Conventional medications such as opioids and antidepressants have poor efficacy for managing chronic pain and possess serious central side-effects, such as altered mental state, addiction and life-threatening respiratory depression.
Dr. Andrea Nackley is committed to improving pain management strategies for patients with CPPCs and, together with her team of investigators, she has identified a novel target for analgesic development. In clinical studies, they determined that patients with CPPCs have increased levels of catecholamines alongside reduced levels of the enzyme catechol-O-methyltransferase (COMT) that metabolizes catecholamines. Consistent with clinical findings, the Nackley lab has shown that pharmacologic inhibition or genetic knockdown of COMT in rodents produces pain at multiple body sites. Catecholamines are produced by numerous cell types in peripheral, spinal and central tissues where they may bind different types of adrenergic or dopaminergic receptors to produce pain. After an extensive pharmacologic campaign, together with site-of-action studies, Nackley and her team have determined that catecholamine activation of adrenergic receptor beta-3 (Adrb3) in peripheral tissues is a key driver of primary pain.
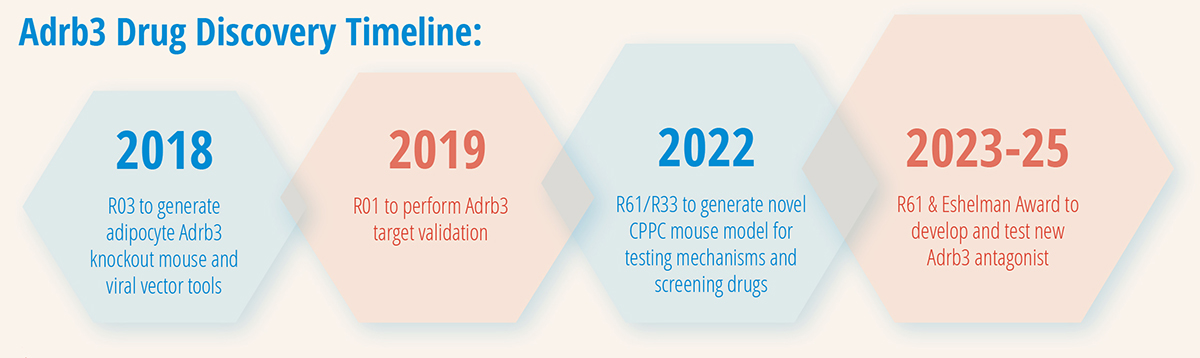

01/15/2018 - 12/31/2020
NIH/NINDS R03 “Defining the Role of Adipocyte Adrb3 in Chronic Pain”
Total Award: $160,000
09/15/2019 - 06/30/2024
NIH/NINDS R01 “Defining the Role of Peripheral Adrb3 in Chronic Pain and Inflammation”
Total Award: $2,623,436
04/01/2022 - 03/31/2025
NIH/NINDS R61/R33 “A Novel Clinically-Relevant Mouse Model of Chronic Overlapping Pain Conditions for Screening Analgesics”
Total Award: $1,150,797
04/01/2023 - 3/31/2025
Eshelman Institute for Innovation Grant “Peripherally-Targeted Adrb3 Antagonists for the Treatment of Pain”
Total Award: $765,444
09/08/2023 - 08/31/2025
NIH/NINDS R61 “Development of Adrb3 Antagonists for the Treatment of Pain”
Total Award: $1,846,202
Adrb3 is predominantly expressed on adipocytes in adipose tissue, where it regulates energy storage and temperature. New data in the Nackley lab suggest that adipocyte Adrb3 also plays a critical role in pain by promoting the synthesis and release of catecholamines, inflammatory mediators and microRNAs that regulate expression of pain-relevant genes. Adrb3 and its downstream effectors promote both nociception, characterized by increased activity of pain-sensing neurons, and neuroinflammation, characterized by increased activity of microglia and astrocytes in regions of the spinal cord and brain that transmit and modulate pain. To directly test the role of adipocyte Adrb3 in pain, Nackley and her team generated mice with conditional Adrb3 knockout only in adipocytes. Remarkably, adipocyte Adrb3 knockout mice do not develop multisite body pain, inflammation or neuroinflammation. Thus, Adrb3 is an exciting new target for development of peripherally-restricted therapies with the potential to overcome the limitations of specificity and central side-effects associated with current treatments.
To translate these research findings into improved analgesic strategies for patients with CPPCs, Nackley recently made two significant advances. First, through an NIH HEAL (Helping to End Addiction Long-term®) Initiative R61 award, she assembled a multidisciplinary team of experts in drug discovery and development to synthesize new Adrb3 antagonists. Inhibition as a therapeutic indication of Adrb3, especially in the context of pain, represents a significant innovation as no Adrb3 antagonists are currently approved for clinical use. The objective of this project is to synthesize, screen and test new potent, selective and peripherally-restricted Adrb3 antagonists.
Second, through an NIH IGNITE (Innovation Grants to Nurture Initial Translational Efforts) R61/R33 award, the Nackley lab developed a novel mouse model of CPPCs that integrates clinically-relevant factors that can be used for testing Adrb3 antagonists as well as other pharmacologic and non-pharmacologic analgesics. Current animal models of CPPCs typically induce pain using chemical irritants and infectious agents that are not implicated in the biological basis of disease. Further, current models typically focus on site-specific pain. Their new model integrates clinically-relevant genetic (COMT knockdown) and environmental (stress) factors to generate multi-site body pain and depressive-like behavior lasting more than three months. In line with the female predominance of CPPCs clinically, the pain and depressive-like behavior in the CPPC mouse model was of greater magnitude and longer duration (>12 months) in females compared to males. Further, they demonstrated the predictive validity of the model as pain was effectively alleviated by an Adrb3 antagonist, but not by drugs that previously failed clinical trials for neuropathic or inflammatory pain.
Successful completion of this ongoing research will advance new peripherally-restricted analgesics into human trials with the potential to reduce the use of opioids in clinical practice and positively impact the quality of life for the 100 million patients across the nation alone who suffer from chronic primary pain. BP
PUBLICATION REFERENCES:
- Zhang X, Kanter K, Chen J, Kim S, Wang Y, Adeyemi C, O’Buckley SC, and Nackley AG. Low catechol-O-methyltransferase and stress potentiate functional pain and depressive behavior, especially in female mice. Pain 2020 Feb 161(2):446-458.
- Wang Y, Kim SH, Klein ME, Chen J, Gu E, Smith SB, Bortsov A, Slade GD, Zhang X, and Nackley AG. A novel mouse model of chronic primary pain conditions that integrates clinically relevant genetic and environmental factors. Science Translational Medicine. 2024 Apr 10;16(742).
Exploring Microbiota-Immune Interactions in Surgery and Critical Illness
Mara Serbanescu, MD
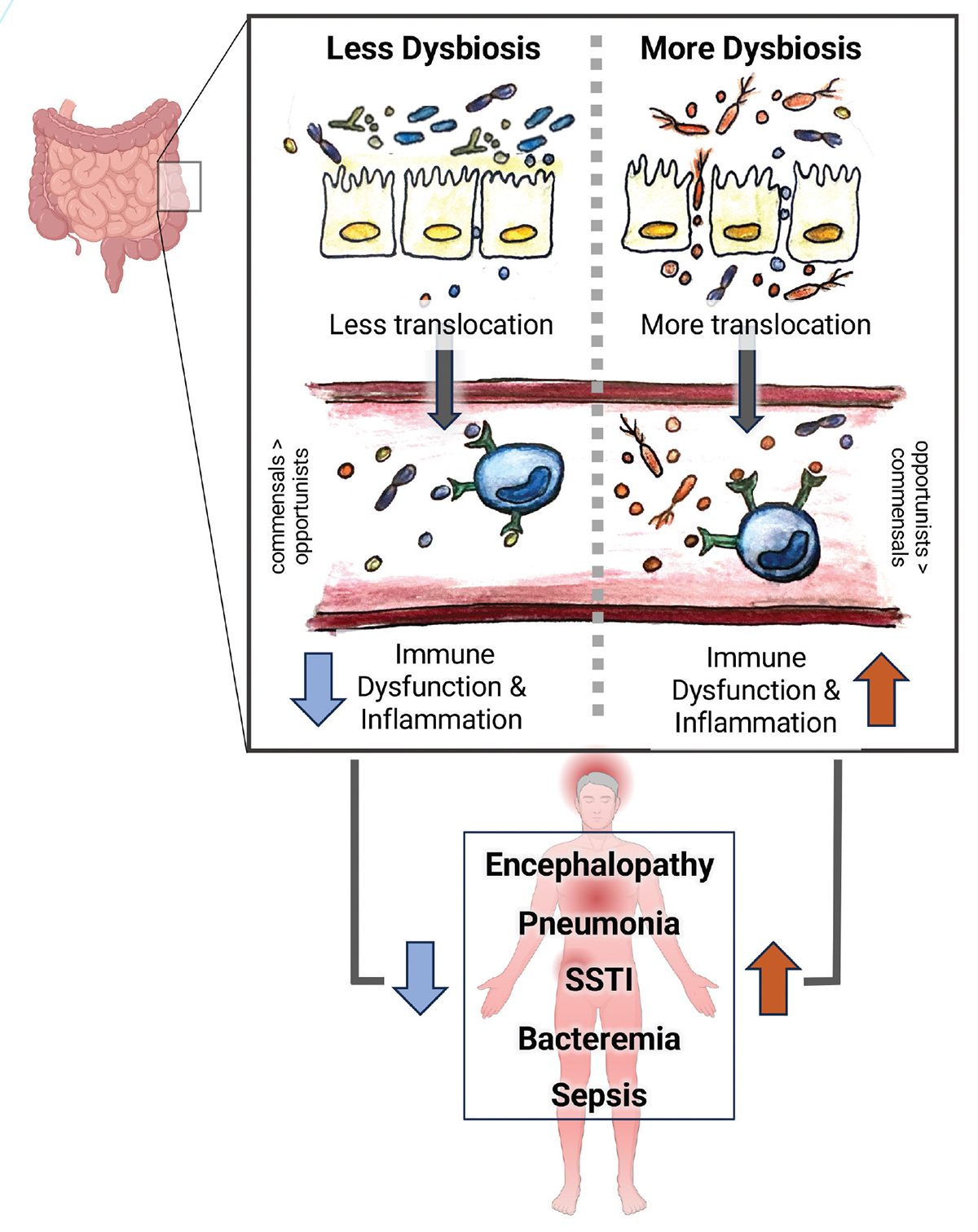
The community of microbes residing in the gastrointestinal tract (i.e., the gut microbiota) plays a crucial role in various processes essential to recovery after surgery and critical illness. Through direct signaling and indirect effects mediated by secreted molecules, the microbiota communicate with local and systemic host immune cells. The types of microbes present (composition) can thus influence immune cell activation, release of inflammatory cytokines, neuronal machinery, and metabolic responses. Following surgery or during critical illness, patients demonstrate a profound loss of beneficial commensals and a relative increase in opportunistic or pathogenic species. In animal models, these changes can lead to complications, such as inflammation-induced organ dysfunction, infection and sepsis. However, if, and how, certain gut microbial signatures influence immune responses in critically ill and post surgical patients is poorly understood.
Dr. Mara Serbanescu, director of the Duke Anesthesiology Microbiome Profiling (Duke AMP) Laboratory, is dedicated to unraveling the complex host-microbiome relationships contributing to complications in critically ill and postoperative patients. Leveraging systems-level approaches integrating microbial genome community profiling, immune function markers, metabolomics, and clinical data, as well as ex-vivo techniques, Serbanescu’s lab aims to identify how dynamic changes in microbiota-immune crosstalk modify clinical trajectory and outcomes. Her long-term goal is to uncover modifiable features of the microbiome that can be targeted to improve outcomes after surgery and critical illness.
Microbial translocation has long been suspected to contribute to inflammatory responses in these conditions, though seminal studies relied on growth in culture for microbial identification and shaped the current understanding that microbe-associated molecular patterns (MAMPs) are stochastically released from the gut and indiscriminately promote systemic inflammation. However, recent culture-independent sequencing technologies like 16S ribosomal RNA (rRNA) profiling have since revolutionized the ability to identify microbial components. Emerging data using these techniques suggests that microbial components can be protective or harmful, influencing discrete immune cell activation pathways and functions. Using 16S rRNA profiling in a perioperative mouse model of severe inflammation, Serbanescu previously executed a proof-of-concept study to examine whether the composition of the gut microbiota influences microbial DNA signatures in the blood, as well as immune responses. She found that following an inflammatory insult, both the extent of microbial translocation and the types of blood microbial signatures were directly shaped by patterns in the gut microbiota present before the insult and associated with specific changes in the immune cell landscape. Serbanescu is now aiming to translate her preclinical insights to clinical practice. Building upon her previous work, she aims to identify whether gut and blood microbial patterns in postoperative and critically ill patients underlie inflammation-induced organ dysfunction and development of secondary infections – the leading causes of morbidity and mortality in these populations. To support her novel area of exploration, she has been awarded a two-year Mentored Research Award for her project titled, “Role of Nutrition on Gut Microbes and Translocation After Trauma Laparotomy” by the International Anesthesiology Research Society (IARS), as well as the Duke Physician Strong Start Award.
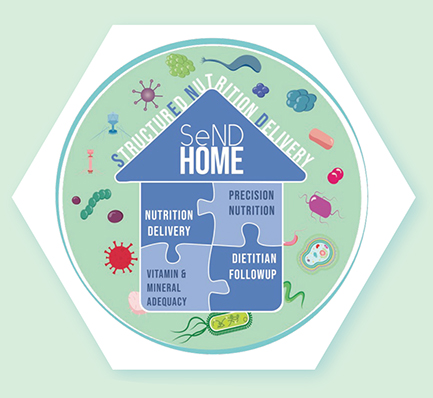
To accomplish her research agenda, Serbanescu has assembled a multidisciplinary team of collaborators, including her mentor, Dr. Paul Wischmeyer, a professor of anesthesiology and surgery renowned for his expertise in perioperative nutrition, and biostatistician Mary Cooter Wright, who has extensive expertise in computational modeling, as well as collaborators from the Duke Microbiome Center, including Dr. Jason Arnold. In her IARS-funded study, Serbanescu is additionally partnering with Duke Surgery’s Dr. Krista Haines and her team, leveraging the SeND Home Trial — a Department of Defense funded randomized controlled trial led by Haines and Wischmeyer. SeND Home (Figure 1) aims to evaluate the effect of an early personalized structured nutrition program that includes early parenteral nutrition on functional outcomes in critically ill patients admitted after severe abdominal trauma. Serbanescu and her team hypothesize that early structured nutrition may also mitigate certain derangements in gut microbial composition and intestinal permeability, thus reducing microbial translocation and inflammation in the days following emergent surgery. To delineate these complex host-microbe relationships, the researchers will use integrated analyses of gut and blood microbial profiles and inflammatory biomarkers from patient samples collected during ICU stay. This study is poised to be the first in trauma to use culture-independent sequencing techniques to investigate gut and blood microbial DNA signatures and will additionally reveal the impact of readily available nutritional interventions on microbial translocation. Findings from this work and her other planned experiments have the potential to revolutionize our understanding of microbiota-immune interactions in surgical and critical care settings and vitally contribute to the development of novel therapies to improve patient care and outcomes. BP
Advancing Pain Omics
Shad Smith, PhD
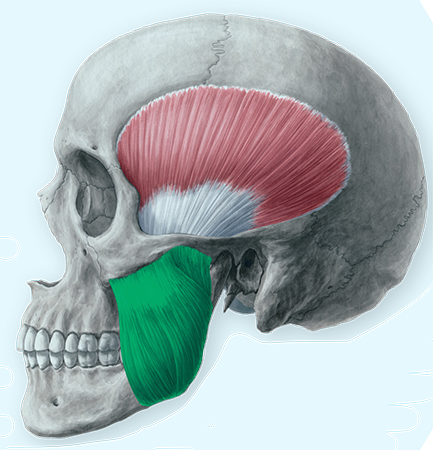
Painful temporomandibular disorders (TMD) are a common set of multifactorial health conditions associated with the temporomandibular joint and surrounding tissues. Estimates suggest that 5% of adults in the United States suffer from TMD pain, making it the leading cause of chronic orofacial pain. There are at present few available mechanism-based tests, and no objective biomarkers, to assist diagnosing or treating TMD. Patients and physicians have therefore struggled with challenges of diagnosis and proper care, as visible signs of trauma or deterioration of the joint are not always present or do not necessarily correspond with the severity of the disorder.
Based on evidence from a 15-year prospective study of TMD, known as the OPPERA study, Dr. Shad Smith’s research approaches TMD as one of a family of “chronic overlapping pain conditions” (COPCs), which arise from a variety of peripheral and central mechanisms that sensitize nociceptors. He has previously identified innate factors that underlie these conditions using large genome screens to identify genes and pathways associated with COPCs. The OPPERA study was also used to identify patterns in predictive characteristics, such as psychological distress and mechanical pain hypersensitivity, that are useful in clustering COPC patients into groups with similar long-term outcomes. Smith believes that identifying clusters of patients with homogeneous features, regardless of their pain diagnosis, could shed light on the underlying mechanisms that are responsible for producing pain.
A recent multiphase UH2/UH3 award funded by the National Institute for Dental and Craniofacial Research (NIDCR) will allow Smith’s team to investigate genetic mechanisms of TMD at a far greater resolution than previously has been possible. New single-cell RNA sequencing technologies (scRNA-seq) are able to identify individual cells and their transcriptional activity, allowing the ability to visualize precisely the cells that are contributing to hypersensitivity and pain. In collaboration with other NIDCR researchers, including Dr. Sunil Kapila of UCLA and Dr. Ryan Tomlinson of Thomas Jefferson University, this project will use scRNA-seq to examine TMJ tissues at cellular resolution for the first time.
FIRST STUDY TO...
... look at masseter muscle in TMD patients
... examine painful masseter muscle tissue using single-cell techniques to discover pathological mechanisms in critical cell populations and characterize their transcriptomic profiles
... conduct simultaneous assessment of multiple tissue types to discover biomarkers correlated with localized pathology in clinically available blood within the same patient
... use these techniques within the context of understanding the origins of TMD pain
A limited understanding of pathological processes in local tissues has hindered the identification of diagnostic, predictive or prognostic biomarkers in more accessible tissues like blood. This project will be the first study to examine painful masseter muscle tissue using single-cell techniques to discover pathological mechanisms in critical cell populations and characterize their transcriptomic profiles. This project will also be the first simultaneous assessment of multiple tissue types to discover biomarkers correlated with localized pathology in clinically available blood within the same patient. The availability of multiple tissue types for analysis is made possible through the assistance of Duke Anesthesiology’s Center for Translational Pain Medicine biorepository, which also under the direction of Smith enrolls COPC patients at the Duke Innovative Pain Therapies (DIPT) clinic and collects specimens and clinical data for research studies. Collaborators in this work include Dr. Daniela Vivaldi, orofacial pain specialist at the DIPT clinic, and other Duke researchers with extensive experience working with scRNA-seq data, Dr. Christopher Donnelly and Dr. Zhicheng “Jason” Ji.
A preliminary phase of this project, currently underway, is a pilot study to develop novel methods of specimen collection and analysis for masseter muscle and blood suitable for high-resolution scRNA-seq techniques. Following development and optimization studies, the team will enroll a cohort of TMD patients and TMD-free controls to identify patterns of phenotypic and molecular expression that distinguish disease-relevant mechanisms. This analysis will also look at clusters of patients with common clinical features to identify specific pathological mechanisms in these subgroups in order to refine diagnostic approaches and identify potential new therapeutic targets.
“Temporomandibular disorder (TMD) significantly restricts essential functions such as speaking and eating, thereby profoundly impacting patients’ connections to their loved ones and even their sense of self. However, the intricate interplay between changes in the nervous and immune systems to produce chronic TMD pain remains ambiguous. Our goal for this study is to gain more insight into these processes, enabling us to target the pathology directly rather than mere symptomatic management.” – DR. SHAD SMITH
Different but complementary approaches will be taken between blood samples and masseter muscle tissue. Circulating cells, known as peripheral blood mononuclear cells (PBMCs), will be analyzed using a method called ATAC-seq to identify not just the immune cell signatures that differentiate health and disease states, but also epigenetic regulatory networks that may help sustain immune dysregulation. In painful muscle tissue drawn from the masseter muscle, a technique called spatial transcriptomics will be used to localize cell populations which are altered in abundance in TMD patients versus healthy controls and determine whether these alterations correlate with nerve fiber density.
Notably, this will be the first time these techniques have been used within the context of understanding the origins of TMD pain. The proposed experiments are expected to yield novel insights into the cellular and molecular drivers of TMD pain, and their successful completion will establish a pipeline that can be generalized to other COPC disease cohorts for deeper exploration of pain mechanisms. BP
